epizootic is an extension to poems, a
spatially-explicit, process-explicit, pattern-oriented framework for
modeling population dynamics. This extension adds functionality for
modeling disease dynamics in wildlife. It also adds capability for
seasonality and for unique dispersal dynamics for each life cycle
stage.
You can install the latest release on CRAN with:
install.packages("epizootic")You can install the latest version of epizootic from GitHub with:
# install.packages("devtools")
install.packages("poems")
devtools::install_github("viralemergence/epizootic")Because epizootic is an extension to poems,
it is necessary to install poems first.
poems and epizootic run on R6 classes. R
is primarily a functional programming language in which the
primary units of programming are expressions and functions. Here we use
R6 to create an object-oriented framework inside of R. R6
classes such as DiseaseModel and
SimulationHandler are used to store model attributes, check
them for consistency, pass them to parallel sessions for simulation, and
gather results and errors.
Here is the initial state of an idealized theoretical disease scenario, following a SIR disease model with three life cycle stages: juvenile, yearling, and adult.
library(poems)
library(purrr)
library(epizootic)
example_region <- Region$new(coordinates = data.frame(x = rep(seq(177.01, 177.05, 0.01), 5),
y = rep(seq(-18.01, -18.05, -0.01), each = 5)))
initial_abundance <- c(c(5000, 5000, 5000, 1, 0, 0, 0, 0, 0),
rep(c(5000, 5000, 5000, 0, 0, 0, 0, 0, 0), 24)) |>
matrix(nrow = 9)
example_region$raster_from_values(initial_abundance[3,]) |>
raster::plot(main = "Susceptible Adults")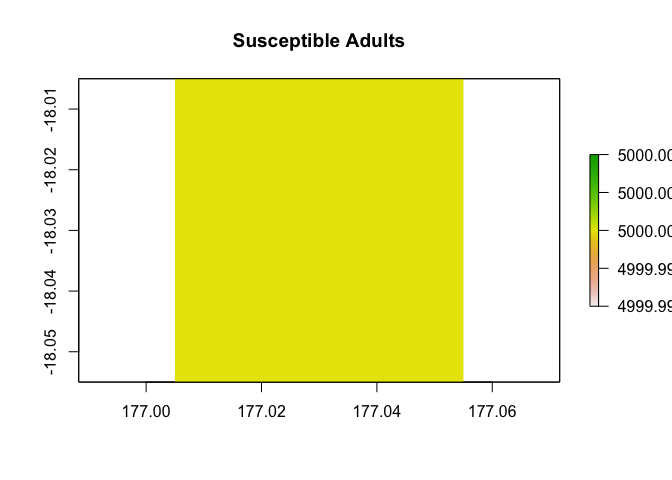
example_region$raster_from_values(initial_abundance[4,]) |>
raster::plot(main = "Infected Juveniles")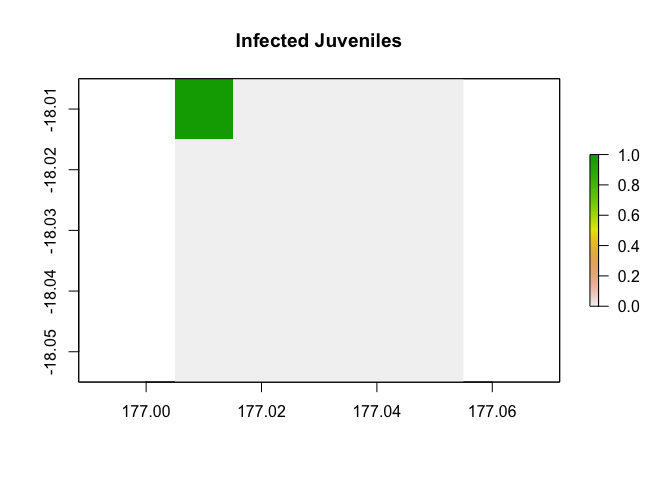
Here I create a DiseaseModel object, which stores inputs
for disease simulations and checks them for consistency and
completeness.
model_inputs <- DiseaseModel$new(
time_steps = 10,
seasons = 2,
populations = 25,
stages = 3,
compartments = 3, # indicates disease compartments
region = example_region,
initial_abundance = initial_abundance,
# Dimensions of carrying_capacity are populations by timesteps
carrying_capacity = matrix(100000, nrow = 25, ncol = 10),
# Indicates length of breeding season in days for each population
breeding_season_length = rep(100, 25),
# One mortality value for each stage and compartment
mortality = c(0.4, 0.2, 0, 0.505, 0.25, 0.105, 0.4, 0.2, 0),
# Indicates that these are seasonal mortality values
mortality_unit = 1,
# No reproduction in this simple example
fecundity = 0,
fecundity_unit = 1,
fecundity_mask = rep(0, 9),
# Transmission rates from infected individuals, one for each stage
transmission = c(0.00002, 0.00001, 7.84e-06),
# Indicates that these are daily transmission rates
transmission_unit = 0,
# Indicates that all stages in the first compartment, S, can be infected
transmission_mask = c(1, 1, 1, 0, 0, 0, 0, 0, 0),
recovery = c(0.05714286, 0.06, 0.1),
recovery_unit = 0,
# Indicates that all stages in the second compartment, I, can recover
recovery_mask = c(0, 0, 0, 1, 1, 1, 0, 0, 0),
season_functions = list(sir_model_summer, NULL),
dispersal = list(disperser),
simulation_order = list(c("transition", "season_functions", "results"),
c("dispersal", "results")),
verbose = F
)
model_inputs$is_complete()
#> [1] TRUE
model_inputs$is_consistent()
#> [1] TRUEThe core simulation engine of epizootic is the function
disease_simulator, which simulates spatially explicit
disease dynamics in populations. Here I show the results from the
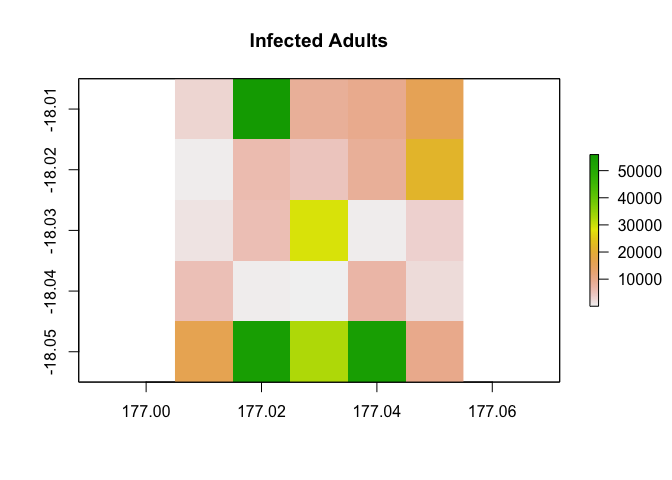
non-breeding season in the tenth year of the simulation.
results <- disease_simulator(model_inputs)
results$abundance_segments$stage_3_compartment_1[,10,2] |>
example_region$raster_from_values() |>
raster::plot(main = "Susceptible Adults")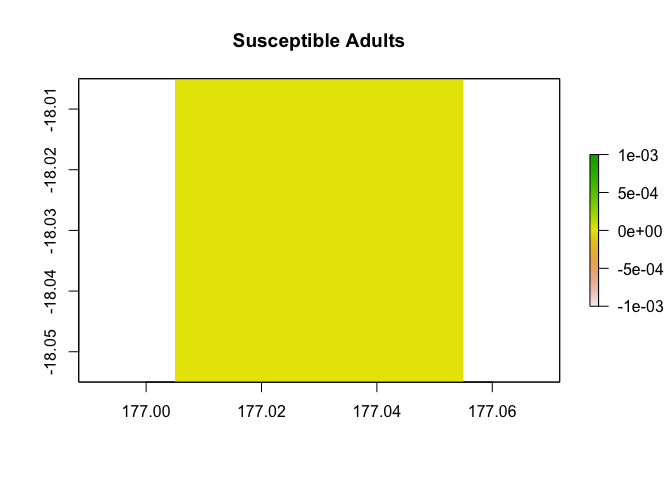
results$abundance_segments$stage_3_compartment_2[,10,2] |>
example_region$raster_from_values() |>
raster::plot(main = "Infected Adults")