

The xrnet R package is an extension of regularized regression (i.e. ridge regression) that enables the incorporation of external data that may be informative for the effects of predictors on an outcome of interest. Let \(y\) be an n-dimensional observed outcome vector, \(X\) be a set of p potential predictors observed on the n observations, and \(Z\) be a set of q external features available for the p predictors. Our model builds off the standard two-level hierarchical regression model,
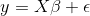
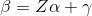
but allows regularization of both the predictors and the external features, where beta is the vector of coefficients describing the association of each predictor with the outcome and alpha is the vector of coefficients describing the association of each external feature with the predictor coefficients, beta. As an example, assume that the outcome is continuous and that we want to apply a ridge penalty to the predictors and lasso penalty to the external features. We minimize the following objective function (ignoring intercept terms):
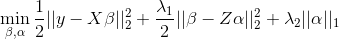
Note that our model allows for the predictor coefficients, beta, to shrink towards potentially informative values based on the matrix \(Z\). In the event the external data is not informative, we can shrink alpha towards zero, returning back to a standard regularized regression. To efficiently fit the model, we rewrite this convex optimization with the variable substitution \(gamma = beta - Z * alpha\). The problem is then solved as a standard regularized regression in which we allow the penalty value and type (ridge / lasso) to be variable-specific:
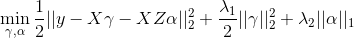
This package extends the coordinate descent algorithm of Friedman et al. 2010 (used in the R package glmnet) to allow for this variable-specific generalization and to fit the model described above. Currently, we allow for continuous and binary outcomes, but plan to extend to other outcomes (i.e. survival) in the next release.
install.packages("xrnet")# Master branch
devtools::install_github("USCbiostats/xrnet")As an example of how you might use xrnet, we have provided a small set of simulated external data variables (ext), predictors (x), and a continuous outcome variable (y). First, load the package and the example data:
library(xrnet)
data(GaussianExample)To fit a linear hierarchical regularized regression model, use the
main xrnet function. At a minimum, you should specify the
predictor matrix x, outcome variable y, and
family (outcome distribution). The external
option allows you to incorporate external data in the regularized
regression model. If you do not include external data, a standard
regularized regression model will be fit. By default, a lasso penalty is
applied to both the predictors and the external data.
xrnet_model <- xrnet(
x = x_linear,
y = y_linear,
external = ext_linear,
family = "gaussian"
)To modify the regularization terms and penalty path associated with
the predictors or external data, you can use the
define_penalty function. This function allows you to
configure the following regularization attributes:
quantile
used to specify quantile, not currently implemented)As an example, we may want to apply a ridge penalty to the x
variables and a lasso penalty to the external data variables. In
addition, we may want to have 30 penalty values computed for the
regularization path associated with both x and external. We modify our
model call to xrnet follows.
penalty_main is used to specify the regularization for
the x variablespenalty_external is used to specify the regularization
for the external variablesxrnet_model <- xrnet(
x = x_linear,
y = y_linear,
external = ext_linear,
family = "gaussian",
penalty_main = define_penalty(0, num_penalty = 30),
penalty_external = define_penalty(1, num_penalty = 30)
)Helper functions are also available to define the available penalty
types (define_lasso, define_ridge, and
define_enet). The example below exemplifies fitting a
standard ridge regression model with 100 penalty values by using the
define_ridge helper function. As mentioned previously, a
standard regularized regression is fit if no external data is
provided.
xrnet_model <- xrnet(
x = x_linear,
y = y_linear,
family = "gaussian",
penalty_main = define_ridge(100)
)In general, we need a method to determine the penalty values that
produce the optimal out-of-sample prediction. We provide a simple
two-dimensional grid search that uses k-fold cross-validation to
determine the optimal values for the penalties. The cross-validation
function tune_xrnet is used as follows.
cv_xrnet <- tune_xrnet(
x = x_linear,
y = y_linear,
external = ext_linear,
family = "gaussian",
penalty_main = define_ridge(),
penalty_external = define_lasso()
)To visualize the results of the cross-validation we provide a contour
plot of the mean cross-validation error across the grid of penalties
with the plot function.
plot(cv_xrnet)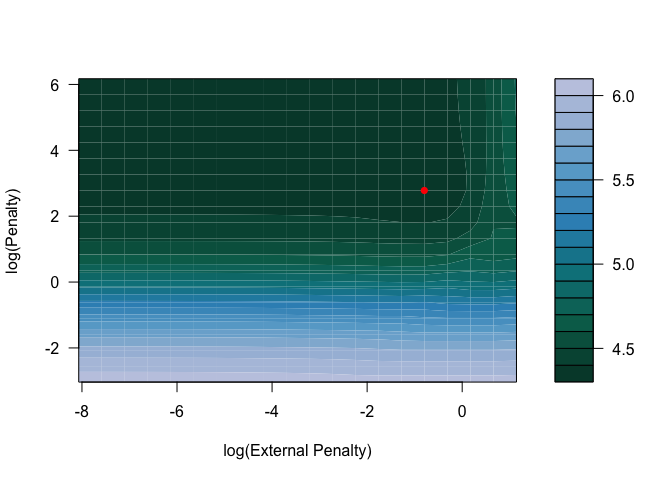
Cross-validation error curves can also be generated with
plot by fixing the value of either the penalty on
x or the external penalty on external. By
default, either penalty defaults the optimal penalty on x
or external.
plot(cv_xrnet, p = "opt")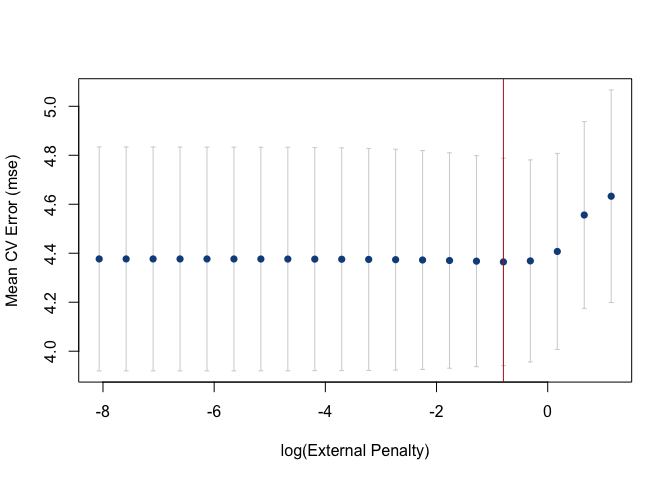
The predict function can be used to predict responses
and to obtain the coefficient estimates at the optimal penalty
combination (the default) or any other penalty combination that is
within the penalty path(s). coef is a another help function
that can be used to return the coefficients for a combination of penalty
values as well.
predy <- predict(cv_xrnet, newdata = x_linear)
estimates <- coef(cv_xrnet)As an example of using bigmemory with
xrnet, we have a provided a ASCII file,
x_linear.txt, that contains the data for x.
The bigmemory function read.big.matrix() can
be used to create a big.matrix version of this file. The
ASCII file is located under inst/extdata in this repository
and is also included when you install the R package. To access the file
in the R package, use
system.file("extdata", "x_linear.txt", package = "xrnet")
as shown in the example below.
x_big <- bigmemory::read.big.matrix(system.file("extdata", "x_linear.txt", package = "xrnet"), type = "double")We can now fit a ridge regression model with the
big.matrix version of the data and verify that we get the
same estimates:
xrnet_model_big <- xrnet(
x = x_big,
y = y_linear,
family = "gaussian",
penalty_main = define_ridge(100)
)
all.equal(xrnet_model$beta0, xrnet_model_big$beta0)
#> [1] TRUEall.equal(xrnet_model$betas, xrnet_model_big$betas)
#> [1] TRUEall.equal(xrnet_model$alphas, xrnet_model_big$alphas)
#> [1] TRUETo report a bug, ask a question, or propose a feature, create a new issue here. This project is released with the following Contributor Code of Conduct. If you would like to contribute, please abide by its terms.
Supported by National Cancer Institute Grant #1P01CA196596.